Network-enhanced feature selection as an enabler of machine learning for eDNA-based ecological assessments of deep-sea ecosystems
Doctoral Researcher:
Hannes Gelbhardt, AWI and Constructor Bremen, hannes.gelbhardt@awi.de
Supervisors:
- Dr. Christina Bienhold, AWI & MPI-MM
- Prof. Marc-Thorsten Hütt, Contructor University Bremen
Location: Bremen
Disciplines: marine microbiology, deep-sea ecology, computational systems biology, machine learning
Keywords: marine eDNA, microbial communities, network analysis, feature selection, explainable AI
Motivation: Marine ecosystems are essential for life on Earth. At the same time, they are threatened by various anthropogenically induced pressures (e.g., climate change, resource exploitation) that act on all parts of the ocean, including deep-sea ecosystems (Boetius & Haeckel, 2018; Levin et al., 2019; Sweetman et al., 2017). The deep sea (>200 m depth) covers around 60% of Earth’s surface and represents about 90% of Earth’s habitable space (Smith et al., 2009).
Deep-sea ecosystems host highly diverse biological communities (Paulus, 2021) and provide key ecosystem functions and services (Thurber et al., 2014). A better understanding of the complex interplay between environmental drivers and biological communities is urgently needed to identify environmental impacts, develop future scenarios and provide policy advice.
Microbial communities (bacteria, archaea, and microbial eukaryotes) are key players in deep-sea ecosystems, detectable via environmental DNA (Sunagawa et al., 2020; Goodwin et al., 2017) and may serve as (bio)indicators for ecosystem state and changes. These communities will be the focus of this project. However, the quantity and complexity of eDNA observations necessitate the application and development of advanced analytical tools to fully exploit the data and facilitate the identification of eDNA-based bioindicators for ecological status (Cordier et al., 2018).
Machine learning (ML) and network approaches have contributed significantly to our understanding of complex systems across a wide range of disciplines. While ML algorithms are capable of detecting subtle patterns in the data, the mechanisms, system features and processes leading to these patterns remain often unexplored. In contrast, network approaches strive for a mechanistic understanding that can provide information about these shortcomings. Additionally, both approaches can provide strategies to reduce the high-dimensionality of the underlying datasets.
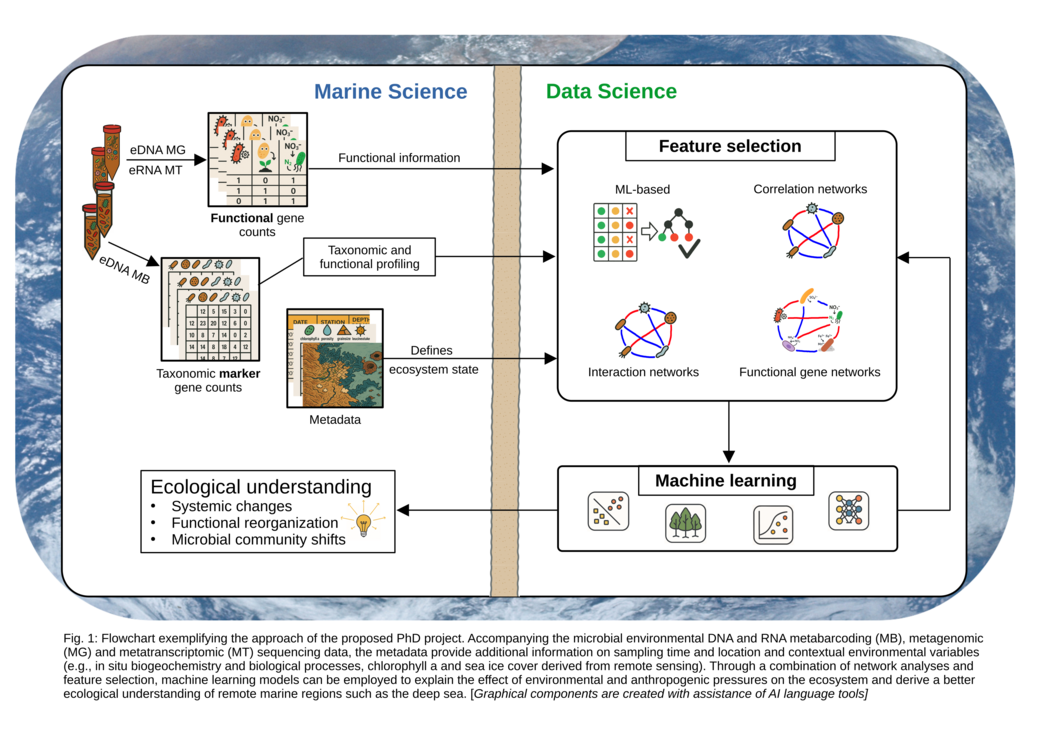
Aim: Different data science approaches will be applied to observations from contrasting deep-sea environments (deep-sea mining test sites, Arctic deep-sea time series) to study how responses in benthic microbial communities reflect environmental impacts and inform on ecosystem state. A key question addresses the best combination of ML and network analysis to maximize sensitivity and specificity for classification while keeping explainability for a systemic understanding. This approach facilitates ecological analyses including the identification of indicator taxa and functions to detect ecosystem change and predict ecosystem characteristics in future scenarios.
The project will develop a framework to identify biological modules/networks, functions and bioindicators to fully explore diverse ecosystem observations collected with utmost efforts and costs within and beyond Helmholtz. Our case studies addressing ecosystem responses to anthropogenic drivers exhibit common shortcomings with sparse observations and countless features/species, and a level of complexity that challenges traditional approaches. To cope with this, a dialog between ML and network science is implemented that allows for (1) network-based feature selection and (2) explainable ML through network interpretation of feature importance.
Objectives:
1.) Develop and implement a combination of ML and network algorithms at the interplay of classification performance and explainability
2.) Employ (network-based) feature selection to improve performance and identify possible bioindicators
3.) Study how responses in benthic microbial communities reflect environmental impacts and inform on ecosystem state
References:
- Boetius, A., Haeckel, M., 2018. Mind the seafloor. Science 359, 34–36. https://doi.org/10.1126/science.aap7301
- Cordier, T., Forster, D., Dufresne, Y., Martins, C.I.M., Stoeck, T., Pawlowski, J., 2018. Supervised machine learning outperforms taxonomy‐based environmental DNA metabarcoding applied to biomonitoring. Molecular Ecology Resources 18, 1381–1391. https://doi.org/10.1111/1755-0998.12926
- Goodwin, K.D., Thompson, L.R., Duarte, B., Kahlke, T., Thompson, A.R., Marques, J.C., Caçador, I., 2017. DNA Sequencing as a Tool to Monitor Marine Ecological Status. Front. Mar. Sci. 4, 107. https://doi.org/10.3389/fmars.2017.00107
- Levin, L.A., Bett, B.J., Gates, A.R., Heimbach, P., Howe, B.M., Janssen, F., McCurdy, A., Ruhl, H.A., Snelgrove, P., Stocks, K.I., Bailey, D., Baumann-Pickering, S., Beaverson, C., Benfield, M.C., Booth, D.J., Carreiro-Silva, M., Colaço, A., Eblé, M.C., Fowler, A.M., Gjerde, K.M., Jones, D.O.B., Katsumata, K., Kelley, D., Le Bris, N., Leonardi, A.P., Lejzerowicz, F., Macreadie, P.I., McLean, D., Meitz, F., Morato, T., Netburn, A., Pawlowski, J., Smith, C.R., Sun, S., Uchida, H., Vardaro, M.F., Venkatesan, R., Weller, R.A., 2019. Global Observing Needs in the Deep Ocean. Front. Mar. Sci. 6, 241. https://doi.org/10.3389/fmars.2019.00241
- Paulus, E., 2021. Shedding Light on Deep-Sea Biodiversity—A Highly Vulnerable Habitat in the Face of Anthropogenic Change. Front. Mar. Sci. 8, 667048. https://doi.org/10.3389/fmars.2021.667048
- Smith, K.L., Ruhl, H.A., Bett, B.J., Billett, D.S.M., Lampitt, R.S., Kaufmann, R.S., 2009. Climate, carbon cycling, and deep-ocean ecosystems. Proc. Natl. Acad. Sci. U.S.A. 106, 19211–19218.
https://doi.org/10.1073/pnas.0908322106 - Sunagawa, S., Acinas, S.G., Bork, P., Bowler, C., Tara Oceans Coordinators, Acinas, S.G., Babin, M., Bork, P., Boss, E., Bowler, C., Cochrane, G., De Vargas, C., Follows, M., Gorsky, G., Grimsley, N., Guidi, L., Hingamp, P., Iudicone, D., Jaillon, O., Kandels, S., Karp-Boss, L., Karsenti, E., Lescot, M., Not, F., Ogata, H., Pesant, Stéphane, Poulton, N., Raes, J., Sardet, C., Sieracki, M., Speich, S., Stemmann, L., Sullivan, M.B., Sunagawa, S., Wincker, P., Eveillard, D., Gorsky, G., Guidi, L., Iudicone, D., Karsenti, E., Lombard, F., Ogata, H., Pesant, Stephane, Sullivan, M.B., Wincker, P., De Vargas, C., 2020. Tara Oceans: towards global ocean ecosystems biology. Nat Rev Microbiol 18, 428–445. https://doi.org/10.1038/s41579-020-0364-5
- Sweetman, A.K., Thurber, A.R., Smith, C.R., Levin, L.A., Mora, C., Wei, C.-L., Gooday, A.J., Jones, D.O.B., Rex, M., Yasuhara, M., Ingels, J., Ruhl, H.A., Frieder, C.A., Danovaro, R., Würzberg, L., Baco, A., Grupe, B.M., Pasulka, A., Meyer, K.S., Dunlop, K.M., Henry, L.-A., Roberts, J.M., 2017. Major impacts of climate change on deep-sea benthic ecosystems. Elementa: Science of the Anthropocene 5, 4. https://doi.org/10.1525/elementa.203
- Thurber, A.R., Sweetman, A.K., Narayanaswamy, B.E., Jones, D.O.B., Ingels, J., Hansman, R.L., 2014.
Ecosystem function and services provided by the deep sea. Biogeosciences 11, 3941–3963.
doi.org/10.5194/bg-11-3941-2014